CeBER Publish in Environmental Science & Technology
ENVIRONMENTAL SCIENCE & TECHNOLOGY
51 (5) 2944–2953
"Genome-Resolved Meta-Omics Ties Microbial Dynamics to Process Performance in Biotechnology for Thiocyanate Degradation" - Kantor R.S., Huddy R.J., Iyer R., Thomas B.C., Brown C.T., Anantharaman K., Tringe S., Hettich R.L., Harrison S.T.L. and Banfield J.F.
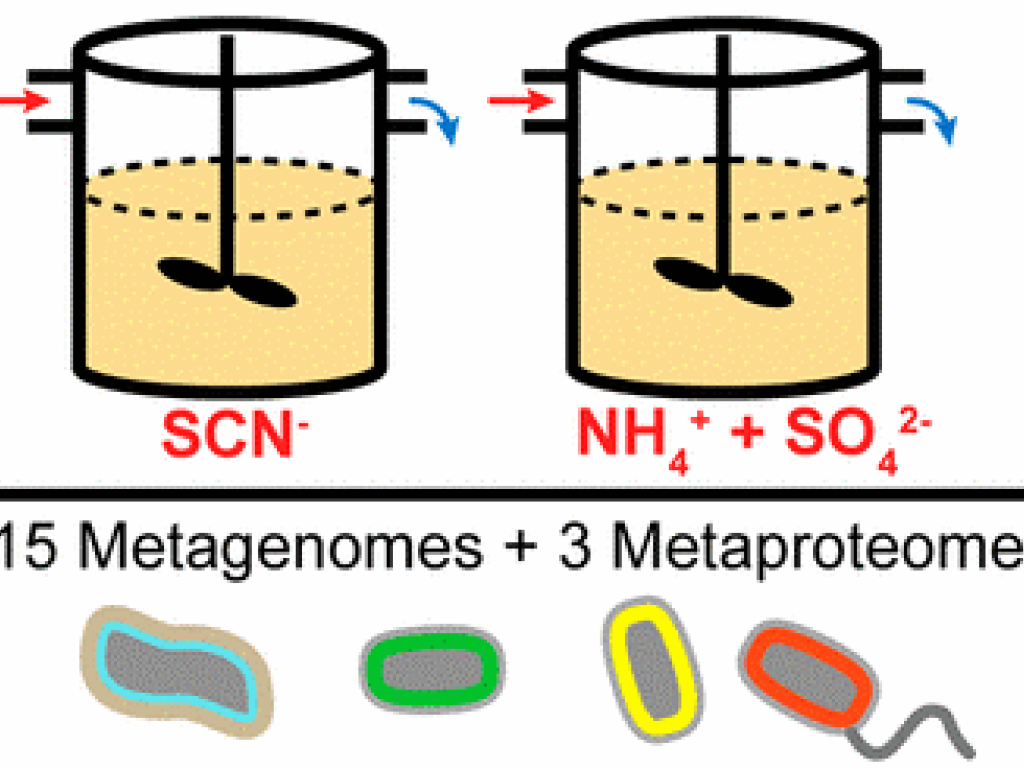
Remediation of industrial wastewater is important for preventing environmental contamination and enabling water reuse. Biological treatment for one industrial contaminant, thiocyanate (SCN–), relies upon microbial hydrolysis, but this process is sensitive to high loadings. To examine the activity and stability of a microbial community over increasing SCN– loadings, we established and operated a continuous-flow bioreactor fed increasing loadings of SCN–. A second reactor was fed ammonium sulfate to mimic breakdown products of SCN–. Biomass was sampled from both reactors for metagenomics and metaproteomics, yielding a set of genomes for 144 bacteria and one rotifer that constituted the abundant community in both reactors. We analyzed the metabolic potential and temporal dynamics of these organisms across the increasing loadings. In the SCN– reactor, Thiobacillus strains capable of SCN– degradation were highly abundant, whereas the ammonium sulfate reactor contained nitrifiers and heterotrophs capable of nitrate reduction. Key organisms in the SCN– reactor expressed proteins involved in SCN– degradation, sulfur oxidation, carbon fixation, and nitrogen removal. Lower performance at higher loadings was linked to changes in microbial community composition. This work provides an example of how meta-omics can increase our understanding of industrial wastewater treatment and inform iterative process design and development.
http://pubs.acs.org/doi/pdf/10.1021/acs.est.6b04477
