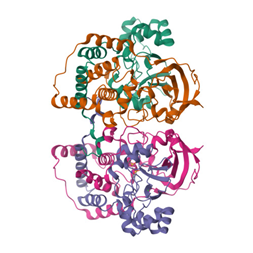
Protein X-ray Crystallography
This technique determines the atomic structure of proteins by analyzing how X-rays scatter when they pass through a crystallized protein. The diffraction pattern is recorded and mathematically transformed into an electron density map, from which the 3D structure of the protein is reconstructed. It relies on the wave properties of X-rays and electron interactions.
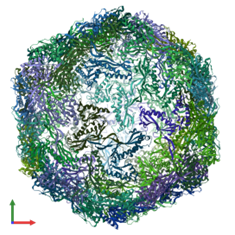
Cryo-EM Single Particle Analysis
In this method, proteins or macromolecules are rapidly frozen in a thin layer of ice, preserving their native structures. A transmission electron microscope (TEM) captures images of individual particles from different orientations. These 2D projections are computationally combined to reconstruct a high-resolution 3D model using principles of Fourier transformation.
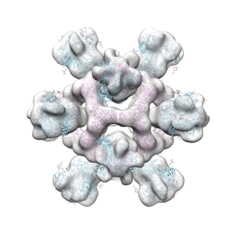
TEM Negative Staining
A biological specimen is coated with an electron-dense stain (e.g., uranyl acetate), which surrounds and enhances the contrast of the sample. Electrons pass through the sample in a TEM, with the stain absorbing or scattering electrons more than the biological material, creating a high-contrast image that reveals overall shape and structure.
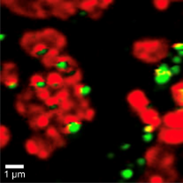
Cryo-EM Tomography
This technique involves freezing a specimen in vitreous ice and capturing multiple tilted projections using a TEM. By computationally reconstructing these images into a 3D volume, the internal structure of cells or macromolecular complexes can be visualized at nanometer resolution, preserving a near-native state without heavy metal stains.

Conventional TEM Tomography
Similar to Cryo-EM tomography, this method collects multiple tilted TEM images of a resin-embedded sample. The images are computationally aligned to generate a 3D reconstruction. Unlike cryo-EM, this method typically requires chemical fixation and heavy metal staining, which can introduce artifacts but provides high-contrast ultrastructural details.
IMAGE
Secondary Electron Scanning Electron Microscopy (SE-SEM)
A focused electron beam scans the surface of a sample, ejecting low-energy secondary electrons, which are detected to form a high-resolution image. SE-SEM provides topographical details of a specimen’s surface and is commonly used for studying morphology at the nanoscale.
IMAGE
Backscattered Electron Scanning Electron Microscopy (BSE-SEM)
In BSE-SEM, high-energy electrons from the SEM beam are elastically scattered back from the sample. The intensity of the backscattered electrons depends on the atomic number of the sample’s elements, providing compositional contrast and allowing differentiation between different materials or structures.
IMAGE
Confocal Raman Microscopy
This technique uses a laser to excite molecular vibrations within a sample, causing Raman scattering. The scattered light is filtered to detect specific vibrational signatures, providing chemical and molecular composition information. The confocal setup enhances spatial resolution by eliminating out-of-focus signals, making it useful for 3D mapping of biochemical distributions in biological samples.